Improving Gene Therapy Viral Vectors Via Capsid Engineering
Nachi Pendse, PhD - January 28, 2022
In this blog post, Dr. Pendse focuses on some of the key directed evolution approaches, strategies, and methodologies that researchers across the field have used to develop novel capsids.
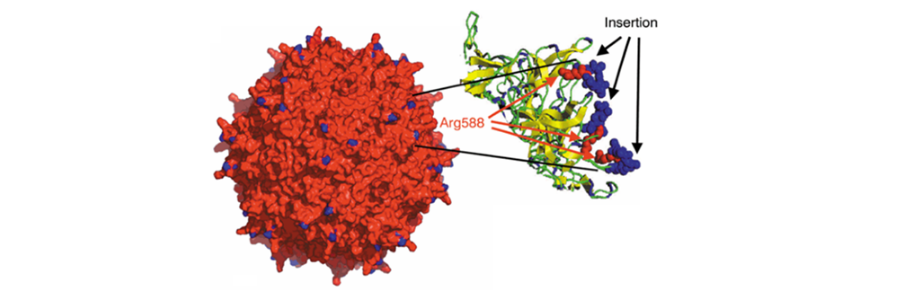
Adeno-associated virus (AAV) vectors are utilized as a key platform for gene therapy and gene delivery for the treatment of a variety of human diseases. The pursuit of finding novel capsids or evolving existing capsid variants to meet clinical safety, efficacy, and potency standards has been a constant expedition in AAV biology for more than 20 years. To advance capsid development, the naturally occurring capsids are further evolved using capsid engineering approaches such as rational design, directed evolution, and in-silico bioinformatics approaches. New strategies for developing novel capsids have evolved with continuous technological progress in methods such as cryo-electron microscopy, multiplexed gene synthesis, next generation sequencing, genetic engineering, AI-based machine learning, and bioinformatic capsid prediction approaches. In this article, I have focused my attention on some of the key directed evolution approaches, strategies, and methodologies that researches across the field have used.
Basic Features of AAVs
AAV belongs to the family Parvoviridae and genus Dependoparvovirus. To complete its life cycle, presence of a helper virus, such as adenovirus- AdV is necessary. AAV is composed of an icosahedral protein capsid and a single-stranded DNA genome of about 4.7 kb which can be present as a sense (plus) or anti-sense (minus) strand. The three capsid protein subunits VP1, VP2, and VP3 comprise the AAV capsids in a ratio of 1:1:10 (VP1:VP2: VP3) for a total of 60 copies. The viral replication needs four rep genes that encodes four proteins: Rep 78, Rep 68, Rep 52, and Rep 40. The AAV genome is flanked by two inverted terminal repeats (ITRs) at the ends that serve as the origins of replication and the packaging signal. The cap gene encodes the three capsid subunits through alternative splicing and translation from different start codons. The assembly activating protein or AAP is another important gene that has been shown to promote virion assembly. This gene is present within the cap coding sequence but in a different reading frame.
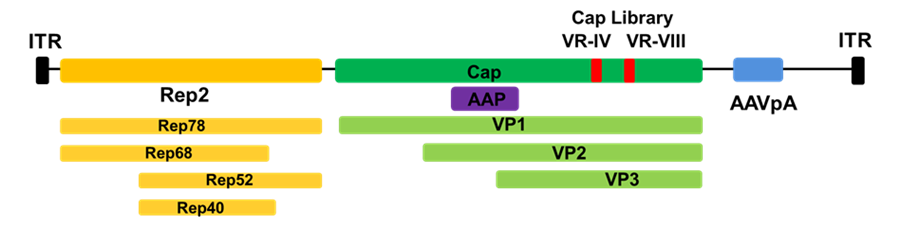
Schematics of AAV2 Genome: AAV has a single stranded linear genome of ~4.7Kb with inverted terminal repeats (ITRs) at the ends. These ITRs flank two viral genes Rep (replication) and Cap (capsid proteins). Cap library insertion sites at VR-IV and VR-VIII marked in red. AAP protein in purple and AAV polyA (pA) signal in blue.
|
Within the coding regions of VP1, VP2, and VP3, there are several variable regions (VR- loop I through IX) which may tolerate the insertion of foreign amino acids (aa) or synthetic peptides. Many of these loops, including VR-IV and VR-VIII, are exposed on the capsid surface and thus are accessible for interaction with external environment or host cell surfaces. From the literature, VR-VIII and VR-IV are the most frequently studied loops for directed evolution approaches. The recent advancements in Cryo-EM have provided insights into the structural and functional features of the capsid residues and variable loops. They have also provided insight into the effects of different aa residues on capsid stability ability to dock on the cell surface receptors, and how antibodies can recognize these capsids. Thus, they provide valuable information on how to manipulate capsid surface residues to target the specific cell types/ tissues- tissue tropism, cross the blood brain barrier (BBB), increase genome packaging capacity, increase transduction efficacy, and may help reduce immunogenicity.
Directed Evolution Approaches to Engineer Capsids
Rational design was one of the first capsid engineering approaches utilized by researchers in the field. In this strategy, known peptide sequence that binds to a cell type specific receptor is grafted. One successful example comes from an early study done by Girod A et al. where researchers introduced the integrin-binding peptide sequences onto the surface of an AAV2 capsid, resulting in an advanced version of AAV2 that could re-target to cells resistant to infection. Later, Warrington KH Jr et al. illustrated that VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. Based on this study, Yang Q et al. and Munch RC et al. utilized the fusion of peptide sequences onto the amino terminus of VP2 such as the single-chain variable fragments – ‘scFvs’ and designed ankyrin repeat proteins – ‘DARPins’ respectively, to confer antibody like recognition to specific cell types. These are just a few examples amongst many from literature that focus on use of rational design strategy.
The key drawback of rational design strategy is the gap in our knowledge related to what specific cell surface residues will interact with capsid surface, inserted peptides, or aa sequence, thereby mediating surface binding, vector internalization, intra-cellular trafficking, and AAV genome expression. Additionally, the rationally designed capsids on one hand may improve the tissue targeting but on the other hand the inserted peptide may affect the stability and fitness of the capsid. To address these issues, researchers have deployed a more sophisticated de-novo approach known as directed evolution. As the name suggests, the idea revolves around mimicking the natural evolution, where the selective pressure is applied on AAV capsid libraries to yield the capsid variants. Directed evolution of capsid libraries does not require preceding knowledge of the molecular/biochemical mechanisms involved in the selection criteria. Using Darwinian principles of “survival of the fittest,” only the best capsid variants are selected with desired traits, such as better tissue specificity, reduced off-targets, reduced immunogenicity, increased transgene expression, increased transduction efficiency, and increased scalability for manufacturing, to name a few.
One of the prominent directed evolution approaches that led to the discovery of PHP.B/eB AAV is known as CREATE (Cre recombination-based AAV targeted evolution) from Viviana Gradinaru’s Lab. This method utilizes the insertion of a randomized 7mer peptide fragment into the Cap9 gene variable region VR loop VIII (aa 588/589) and relies on Cre dependent selective pressure to repair an inverted polyadenylation (pA) sequence designed into the AAV genome.
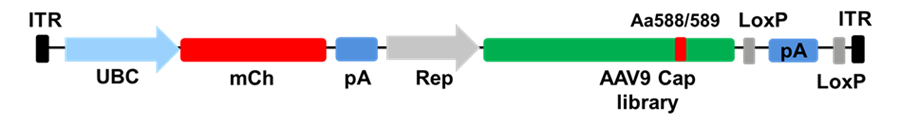
Schematics of CREATE (Cre recombination based AAV targeted evolution) approach: Cell type specific or ubiquitous promotor (UBC) drives the expression of mCherry (mCh), p41/ Rep promotor ensures the expression of AAV Cap9. Cre inverts the pA sites flaked by LoxP sites. pA is in the inverted orientation prior to Cre processing.
Since enzyme Cre-recombinase requires targeting of both strands of DNA, only the capsids that can be efficiently trafficked intracellularly to the nucleus and can convert single stranded genome into double stranded can be processed by Cre-recombinase. PHP.B/eB AAV variant, which showed highly efficient transport over the BBB in C57/BL6 mice. Using Multiplexed M-CREATE platform, the same group further identified CAP-B10 and CAP-B22 that showed promising central nervous system (CNS) delivery in mice and marmosets.

Schematics of TRACER (Tropism Redirection of AAV by Cell-type-specific Expression of RNA) approach. CBA or Synapsis was used as a promotor and AAV9, AAV5, AAV6 and AAV DJ libraries were used. P40 drives the expression of Cap9 gene. AAVpA is marked in blue.
|
While the CREATE approach is novel and resourceful, it is heavily dependent on expression of Ly6A receptors which are unfortunately not found in other mice species,rats, or non-human primates (NHPs), limiting its clinical application via systemic delivery. However, work from Liguore WA et al. and Chatterjee D et al. using PHP.B/eB has successfully shown efficient transduction in the central nervous system in rats and NHPs following intrathecal injections. An alternative approach to address the systemic delivery issue is the Voyager Therapeutics developed TRACER (Tropism Redirection of AAV by Cell-type-specific Expression of RNA) approach. This was tested in mice and NHPs with AAV9 peptide display libraries. The top ten individual variants identified from this study showed up to 400-fold higher brain transduction over AAV9 following systemic administration. Also noteworthy was that capsid TRACER 9P801 showed enhancement in ability to cross BBB and CNS transduction in the NHPs, thus demonstrating the clinical value of this technology.
Discovery of AAV-F and AAV-S capsids, that showed promising transduction to neurons, spinal cord, astrocytes, and muscles (only AAV-S), is based on a new method developed by Casey Maguire’s Lab called iTRANSDUCE.
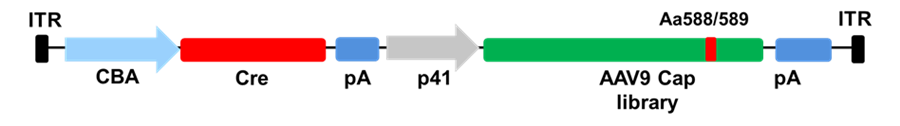
Schematics of iTRANSDUCE approach: CBA promotor drives the expression of Cre- recombinase and p41 promotor drives the Cap9 gene. The peptide display library in variable loop VIII of Cap9 gene is shown in red. pA is marked in blue.
This approach combines a peptide library on the AAV9 capsid with a Cre cassette to enable sensitive detection of transgene expression using a tdTomato reporter system. Like CREATE, iTRANSDUCE also utilizes 7mer randomized peptide insertion in VR-VIII (aa588/589) of Cap9 gene. In two mice strains, C57BL/6 and BALB/c, AAV-F mediated transgene expression in the brain cortex was more than 65-fold higher in astrocytes and 171-fold higher in neurons than the parental AAV9.
With the advancement in multiplex gene synthesis and machine learning tools, the BRAVE (Barcoded Rational AAV Vector Evolution) approach was developed by Tomas Björklund’s Lab.
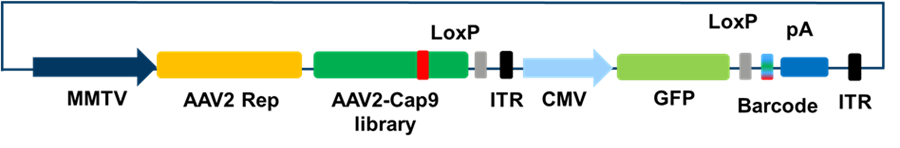
Schematics of BRAVE (Barcoded Rational AAV Vector Evolution) approach: The mouse mammary tumor virus (MMTV) promoter was used to drive the expression of AAV2 Rep and Cap genes. Two inserted LoxP sites allows the paired sequencing of Cap NNK library and Barcode. GFP was used as a reporter and is driven by CMV promotor. The barcoded expression cassette is flanked by two ITRs, and thus packaged inside the formed capsids.
They combined the Cre-approach with sophisticated computational tools to find novel capsid variants by using a sliding window approach-based library of viral proteins or endogenous proteins with desired functions. In this brilliant BRAVE approach, AAV2 Rep/Cap genes are expressed from the same plasmid, but outside the inverted terminal repeats (ITRs), which creates a linkage between capsid structure and Barcode as illustrated above. In the BRAVE approach, each virus particle displays a peptide, derived from a protein, of known function on the AAV capsid surface, and a unique molecular barcode in the packaged genome. From this study, the researchers identified MNM008 and MNM004 AAV2 capsid variant with a unique capacity to reach dopamine neurons in retrograde fashion.
As CREATE, iTRANSDUCE, and BRAVE technologies require Cre-expression or Cre transgene animal models, Nakai H et. al developed Cre- independent approach called TRADE (TRAnscription-dependent Directed Evolution).
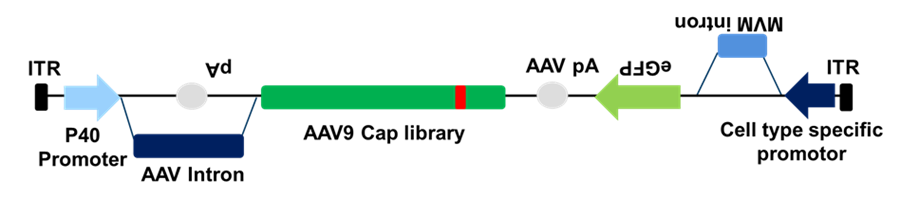
Schematics of TRADE (TRAnscription-dependent Directed Evolution) approach. P40 promotor drives the expression of AAV9 Cap library while cell type specific promotor (in reverse orientation) drives eGFP reporter as shown. TRADE is Cre independent approach and allows selection at mRNA or even at protein level; thus, it can provide greater selective pressure as compared to CREATE.
TRADE is a powerful tool for identifying AAV variants with enhanced transduction at the level of cell specific or mRNA expression which can be applied to a broad range of cell cultures and animal models. Using TRADE approach, the CNS selection yielded AAV-HN capsid variant. Although TRADE approach is Cre-independent and offers mRNA level selection, challenges due to aberrant or cryptic splicing issues of antisense transcript remain to be addressed.
With the recent advances in genome engineering approaches, editing or replacing genetic mutations via CRISPR, base editing, or prime editing holds great promise. However, precise delivery of these components to target tissues remains a key challenge. Pardis Sabeti’s lab recently developed the DELIVER (Directed Evolution of AAV capsids Leveraging In Vivo Expression of transgene RNA) approach that combines the power of capsid engineering and RNA transcript based in vivo selection.

Schematics of DELIVER (Directed Evolution of AAV capsids Leveraging In Vivo Expression of transgene RNA) approach. Ubiquitous CMV or muscle specific CK8 or MHCK7 promoter was used to drive the expression of AAV9 Capsid genes. Between aa 588/596 Cap NNK library was inserted. DELIVER approach based muscle selection resulted in discovery of MyoAAVs.
This criterion allows identification of functional capsid variants in any tissue of interest and any animal model. The DELIVER approach was applied to develop muscle-tropic capsids in mice and NHPs. In comparison to AAV9 and AAVrh74, both of which are currently being used in clinical trials for Duchenne muscular dystrophy, variant MyoAAV from this study showed higher efficacy in NHPs at a much lower dose. This approach also sheds some light on the unique RGD aa sequence which is important for increased MyoAAV interactions with integrin heterodimers in mouse and human cells. In mice, a study was reported by Dirk Grimm’s Lab on a muscle tropic AAVMYO variant containing RGD motif, although no NHPs data is available for this variant. While MyoAAV showed promising effects in NHPs, these parallel efforts resulted in new muscle tropic AAVs for potential future clinical applications.
The approaches presented here, in my opinion, are some of the dynamic and breakthrough studies in the field and have raised the bar quite high for gene therapy. The promising capsids that have emerged from these studies have brought us a step closer to translating gene therapies to patients. Given the competitive landscape of the AAV capsid engineering space and continuous technological advancement in gene editing and gene therapy, there will be many more exciting approaches in pursuit of finding the ideal “designer capsids.”
The views and opinions expressed in this article are those of the author and do not necessarily reflect the official policy or position of Novartis or any of its officers.
Dr. Pendse is senior project lead scientist at Novartis and a member of ASGCT's Communications Committee.